Note
Go to the end to download the full example code
Structural poperties of the MEG-informed parcellations¶
Import the required packages
import os.path as op
import numpy as np
import pickle
import matplotlib.pyplot as plt
import matplotlib.patches as patches
from mne import (read_forward_solution, pick_types_forward,
convert_forward_solution, read_labels_from_annot,
spatial_src_adjacency, read_source_spaces)
from mne.viz import get_brain_class
from mne.datasets import sample
import megicparc
Define input parameters for the flame algorithm running in megicperc
gamma_tot = np.arange(0, 1.01, 0.2)
knn_tot = [10, 20, 30, 40]
theta = 0.05
parc = 'aparc'
sensors_meg = 'grad'
folder_fl = op.join('..', 'data', 'data_mne_sample')
string_target_file = op.join(folder_fl,
'{:s}_flame_grad_k{:d}_gamma{:1.2f}_theta{:1.2f}.pkl')
Load lead-field matrix, source-space and anatomy-based Atlas
data_path = sample.data_path()
subjects_dir = op.join(data_path, 'subjects')
subject = 'sample'
fwd_file = op.join(data_path, 'MEG', subject, 'sample_audvis-meg-eeg-oct-6-fwd.fif')
src_file = op.join(folder_fl, 'source_space_distance-src.fif')
fwd = read_forward_solution(fwd_file)
fwd = pick_types_forward(fwd, meg=sensors_meg, eeg=False,
ref_meg=False, exclude='bads')
fwd = convert_forward_solution(fwd, surf_ori=True, force_fixed=True,
use_cps=True)
src = read_source_spaces(src_file)
fwd['src'] = src
n_vert = fwd['src'][0]['nuse'] + fwd['src'][1]['nuse']
label_lh = read_labels_from_annot(subject=subject, parc=parc, hemi='lh',
subjects_dir=subjects_dir)
label_rh = read_labels_from_annot(subject=subject, parc=parc, hemi='rh',
subjects_dir=subjects_dir)
label = label_lh + label_rh
Reading forward solution from /u/29/sommars1/unix/mne_data/MNE-sample-data/MEG/sample/sample_audvis-meg-eeg-oct-6-fwd.fif...
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
2 source spaces read
Desired named matrix (kind = 3523) not available
Read MEG forward solution (7498 sources, 306 channels, free orientations)
Desired named matrix (kind = 3523) not available
Read EEG forward solution (7498 sources, 60 channels, free orientations)
Forward solutions combined: MEG, EEG
Source spaces transformed to the forward solution coordinate frame
203 out of 366 channels remain after picking
Average patch normals will be employed in the rotation to the local surface coordinates....
Converting to surface-based source orientations...
[done]
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
2 source spaces read
Reading labels from parcellation...
read 34 labels from /u/29/sommars1/unix/mne_data/MNE-sample-data/subjects/sample/label/lh.aparc.annot
Reading labels from parcellation...
read 34 labels from /u/29/sommars1/unix/mne_data/MNE-sample-data/subjects/sample/label/rh.aparc.annot
Initialization
num_roi = np.zeros((np.size(knn_tot), np.size(gamma_tot)))
n_cc = {x : [] for x in \
['k%d_gamma%1.2f'%(knn, gamma) for knn in knn_tot for gamma in gamma_tot]}
coverage = np.zeros((np.size(knn_tot), np.size(gamma_tot)))
Analysis with anatomy-based parcels.
proj_anrois = megicparc.labels_to_array(label, fwd['src'])
# Number of regions
if proj_anrois['outliers'] > 0:
aux_num_roi = len(proj_anrois['parcel']) - 1
else:
aux_num_roi = len(proj_anrois['parcel'])
num_roi_an = aux_num_roi
# Coverage
coverage_an = 100 * (n_vert - proj_anrois['outliers']) / n_vert
Analysis with meg-informed parcels
bmesh_adj_mat = np.array(spatial_src_adjacency(fwd['src']).todense())
for idx_g, gamma in enumerate(gamma_tot):
for idx_k, knn in enumerate(knn_tot):
target_file = string_target_file.format(
subject, knn, gamma, theta)
print('Loading %s'%target_file)
with open(target_file, 'rb') as aux_lf:
flame_data = pickle.load(aux_lf)
# Number of regions
num_roi[idx_k, idx_g] = flame_data['centroids']
# Coverage
coverage[idx_k, idx_g] = \
100 * (n_vert - flame_data['outliers']) / n_vert
# Number of connected components
n_cc['k%d_gamma%1.2f'%(knn, gamma)] += \
[len(megicparc.compute_connected_components(
bmesh_adj_mat, flame_data['parcel'][ir])) \
for ir in range(flame_data['centroids'])]
-- number of adjacent vertices : 8196
/m/home/home2/29/sommars1/data/Documents/megicparc/examples/plot_3_structural_properties.py:86: RuntimeWarning: 8.5% of original source space vertices have been omitted, tri-based adjacency will have holes.
Consider using distance-based adjacency or morphing data to all source space vertices.
bmesh_adj_mat = np.array(spatial_src_adjacency(fwd['src']).todense())
Loading ../data/data_mne_sample/sample_flame_grad_k10_gamma0.00_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k20_gamma0.00_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k30_gamma0.00_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k40_gamma0.00_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k10_gamma0.20_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k20_gamma0.20_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k30_gamma0.20_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k40_gamma0.20_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k10_gamma0.40_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k20_gamma0.40_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k30_gamma0.40_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k40_gamma0.40_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k10_gamma0.60_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k20_gamma0.60_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k30_gamma0.60_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k40_gamma0.60_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k10_gamma0.80_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k20_gamma0.80_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k30_gamma0.80_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k40_gamma0.80_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k10_gamma1.00_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k20_gamma1.00_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k30_gamma1.00_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k40_gamma1.00_theta0.05.pkl
Compute mean and SEM of the number of connected components over the regions of a MEG-informed parcellations
n_cc_ave = np.zeros((np.size(knn_tot), np.size(gamma_tot)))
n_cc_sem = np.zeros((np.size(knn_tot), np.size(gamma_tot)))
for idx_g, gamma in enumerate(gamma_tot):
for idx_k, knn in enumerate(knn_tot):
aux_cc = np.asanyarray(n_cc['k%d_gamma%1.2f'%(knn, gamma)])
n_cc_ave[idx_k, idx_g] = np.mean(aux_cc)
n_cc_sem[idx_k, idx_g] = np.std(aux_cc, ddof=1) / np.sqrt(np.size(aux_cc))
Additional analysis: impact of the anatomical constraints
k_nn_ac = [30]
theta_ac = [0, 0.05]
gamma_ac = np.arange(1, 0.4, -0.2)
tolls = [0.05]
spread = {}
num_roi_constr = {}
for idx_t, theta in enumerate(theta_ac):
for idx_k, knn in enumerate(k_nn_ac):
for idx_g, gamma in enumerate(gamma_ac):
# 6.1. Reload parcellation
target_file = string_target_file.format(subject, knn, gamma,
theta)
print('Loading %s'%target_file)
with open(target_file, 'rb') as aux_lf:
flame_data = pickle.load(aux_lf)
num_roi_constr['k%d_gamma%1.2f_theta%1.2f'%(knn, gamma, theta)] = \
flame_data['centroids']
# 6.2. Compute cotingency matrix
conting_mat = np.array([
np.array([
np.intersect1d(p_fl, p_an).shape[0]
for p_an in proj_anrois['parcel']])
for p_fl in flame_data['parcel'][0:flame_data['centroids']]])
conting_mat = conting_mat / \
np.sum(conting_mat, axis=1)[:, np.newaxis]
# 6.3. Compute spread of flame regions over anatomical
spread['k%d_gamma%1.2f_theta%1.2f'%(knn, gamma, theta)] = \
np.zeros((flame_data['centroids'], np.size(tolls)))
for idx_t, toll in enumerate(tolls):
spread['k%d_gamma%1.2f_theta%1.2f'%(knn, gamma, theta)] [:, idx_t] = \
np.sum(conting_mat > toll, axis=1)
Loading ../data/data_mne_sample/sample_flame_grad_k30_gamma1.00_theta0.00.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k30_gamma0.80_theta0.00.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k30_gamma0.60_theta0.00.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k30_gamma1.00_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k30_gamma0.80_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k30_gamma0.60_theta0.05.pkl
General parameters for plotting
plt.rc('text', usetex=True)
plt.rc('font', family='serif', size=22)
plt.rcParams['axes.grid'] = True
plt.rcParams['lines.linewidth'] = 3
plt.rcParams['errorbar.capsize'] = 3.5
plt.rcParams["figure.figsize"] = [12, 8]
plt.rcParams['patch.force_edgecolor'] = True
colors = np.array([np.array([0, 1, 0]),
np.array([0, 0, 1]),
np.array([1, 0, 0]),
np.array([0, 0.9655, 1])])
Plot 1. Number of regions
f_numreg = plt.figure()
for idx_k, knn in enumerate(knn_tot):
plt.plot(gamma_tot, num_roi[idx_k,], color=colors[idx_k],
label='$k$ = %d'%knn)
plt.plot(gamma_tot, num_roi_an*np.ones(np.size(gamma_tot)),
'k--', label='DK')
plt.legend(loc=[0.68, 0.47], fontsize=18)
plt.xlabel(r'Weight of the spatial distances', fontsize=25)
plt.ylabel(r'Number of parcels', fontsize=25)
plt.ylim(0, 350)
plt.xlim(-0.05, 1.05)
f_numreg.set_size_inches(8.5, 6)
plt.tight_layout(pad=1.5)
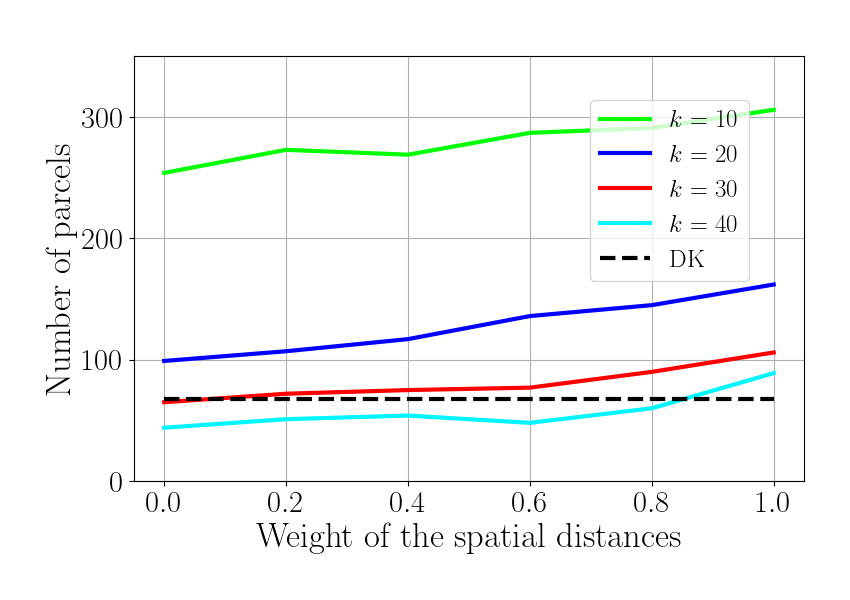
Plot 2. Number of connected components
f_cc = plt.figure()
for idx_k, knn in enumerate(knn_tot):
plt.errorbar(gamma_tot, n_cc_ave[idx_k, :], yerr=n_cc_sem[idx_k, :],
label='$k$ = %d'%knn, color=colors[idx_k])
plt.legend(fontsize=18)
plt.xlabel(r'Weight of the spatial distances', fontsize=25)
plt.ylabel(r'Number connected components', fontsize=25)
plt.ylim(0, 30)
plt.xlim(-0.05, 1.05)
#plt.show()
f_cc.set_size_inches(8.5, 6)
plt.tight_layout(pad=1.5)
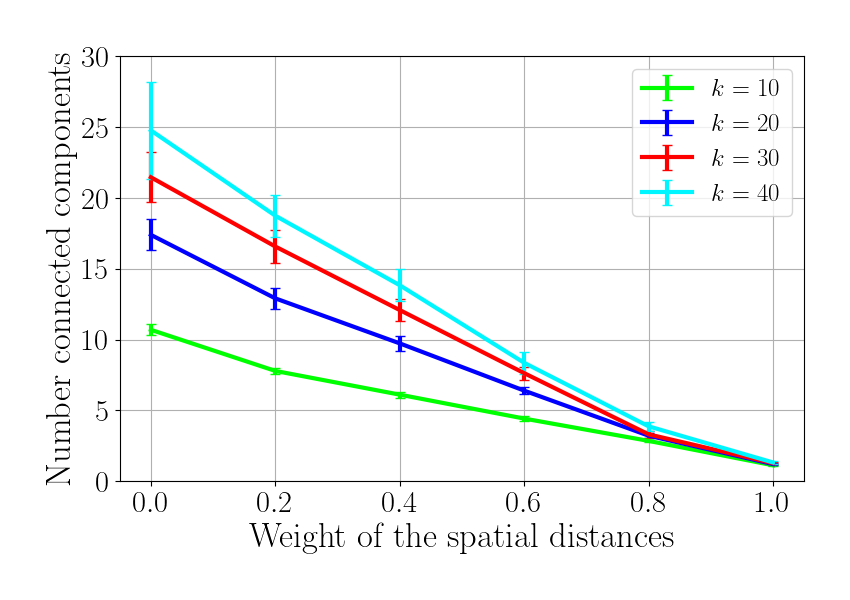
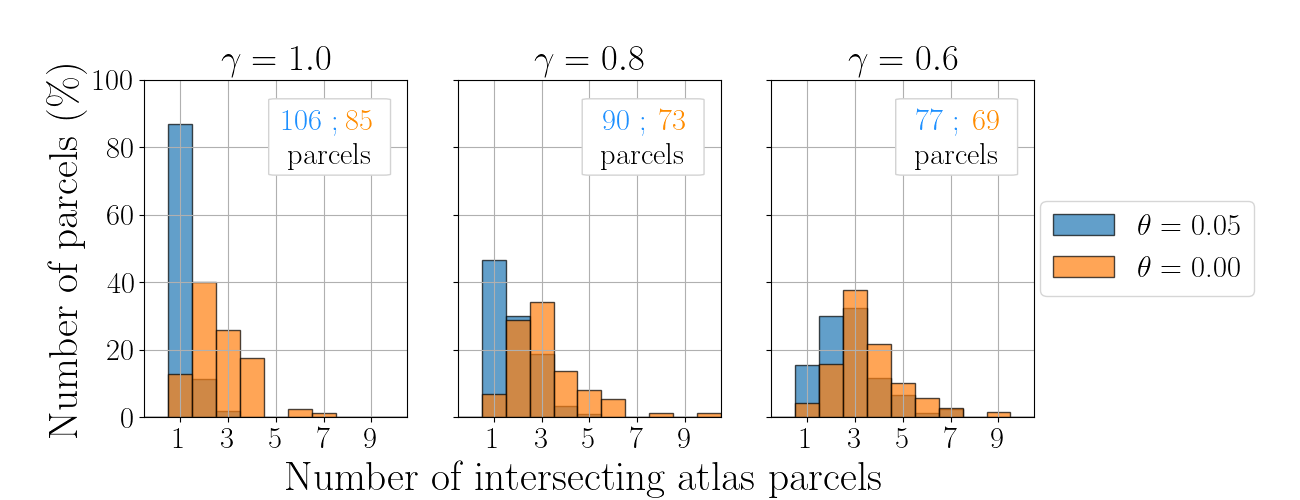
. Plot 3. (Additional) Impact of anatomical constraints
bins = np.arange(-0.5, 69, 1)
alpha = 0.7
knn = k_nn_ac[0]
toll = tolls[0]
fac, axac = plt.subplots(1, np.size(gamma_ac), figsize=[13, 5])
#fac.suptitle('$k =$ %d'%knn)
idx_t = 0
for idx_g, gamma in enumerate(gamma_ac):
aux_th1 = spread['k%d_gamma%1.2f_theta%1.2f'%(knn, gamma, theta_ac[1])][:, idx_t]
axac[idx_g].hist(aux_th1, bins=bins, alpha=alpha,
weights=np.ones_like(aux_th1)/np.shape(aux_th1)[0]*100,
label=r'$\theta = $ %1.2f'%theta_ac[1])
aux_th2 = spread['k%d_gamma%1.2f_theta%1.2f'%(knn, gamma, theta_ac[0])] [:, idx_t]
axac[idx_g].hist(aux_th2, bins=bins, alpha=alpha,
weights=np.ones_like(aux_th2)/np.shape(aux_th2)[0]*100,
label=r'$\theta = $ %1.2f'%theta_ac[0])
axac[idx_g].set_ylim(0, 100)
axac[idx_g].set_xlim(-0.5, 10.5)
#axac[idx_t, idx_g].legend()
if idx_t == 0:
axac[idx_g].set_title(r'$\gamma = %1.1f$ '%(gamma))
axac[idx_g].set_xticks(np.arange(1, 11, 2))
axac[idx_g].text(6.5, 85, '%d ;'%(num_roi_constr['k%d_gamma%1.2f_theta0.05'%(knn, gamma)]),
ha='center', color='dodgerblue', fontweight='bold')
axac[idx_g].text(8.5, 85, '%d'%(num_roi_constr['k%d_gamma%1.2f_theta0.00'%(knn, gamma)]),
ha='center', color='darkorange', fontweight='bold')
axac[idx_g].text(7.2, 75, 'parcels', ha='center')
rect = patches.FancyBboxPatch((5, 72), 4.5, 22,
boxstyle='round', facecolor='white',
edgecolor='lightgray', zorder=2)
axac[idx_g].add_patch(rect)
else:
axac[idx_g].set_xticks(np.arange(1, 11, 2))
if idx_g == 0:
axac[idx_g].set_ylabel(
r'Number of parcels ($\%$)',
fontsize=30)
else:
axac[idx_g].set_yticklabels([])
axac[-1].legend(bbox_to_anchor=(1, 0.5), loc='center left',
borderaxespad=0.2)
fac.text(0.45, 0.02, 'Number of intersecting atlas parcels', ha='center',
fontsize=30)
plt.tight_layout(pad=1.5)
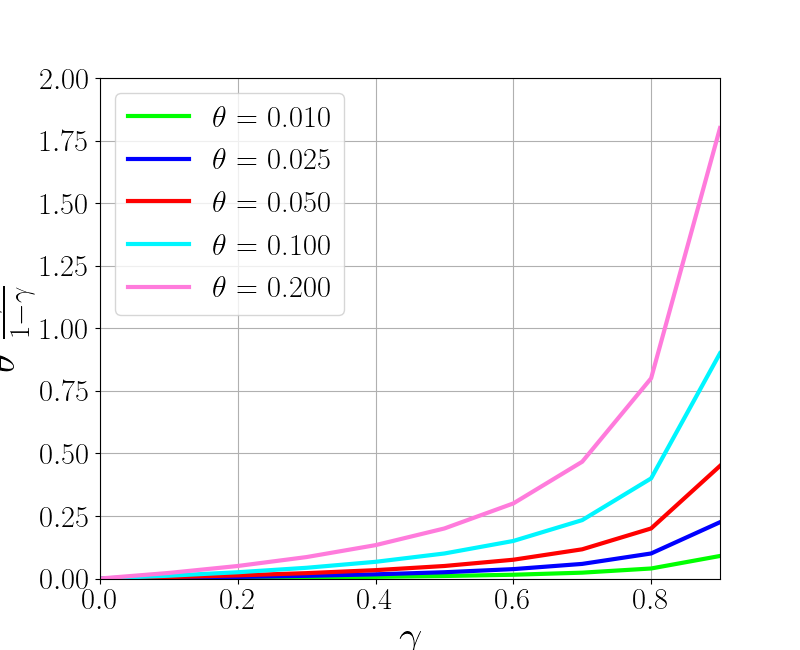
gamma_tplot = np.arange(0, 1, 0.1)
theta_tplot = np.array([0.01, 0.025, 0.05, 0.1, 0.2])
f_tg = np.array([theta * (gamma_tplot / (1-gamma_tplot)) \
for theta in theta_tplot])
colors_tplot = np.array([np.array([0, 1, 0]),
np.array([0, 0, 1]), np.array([1, 0, 0]),
np.array([0, 0.9655, 1]), np.array([1, 0.4828, 0.8621])])
f_tplot = plt.figure()
for idx_t, theta in enumerate(theta_tplot):
plt.plot(gamma_tplot, f_tg[idx_t],
label=r'$\theta$ = %1.3f'%theta, color=colors_tplot[idx_t])
plt.legend()
plt.xlim([0, 0.9])
plt.ylim([0, 2])
plt.xlabel(r'$\gamma$', fontsize=30)
plt.ylabel(r'$\theta$ $\frac{\gamma}{1-\gamma}$', fontsize=30)
f_tplot.set_size_inches(8, 6.5)
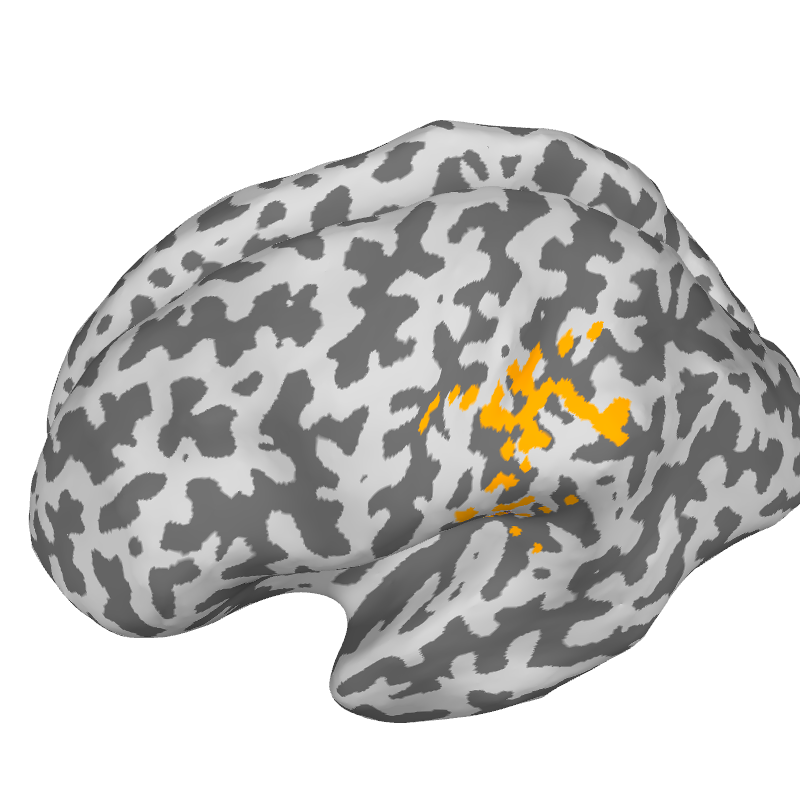
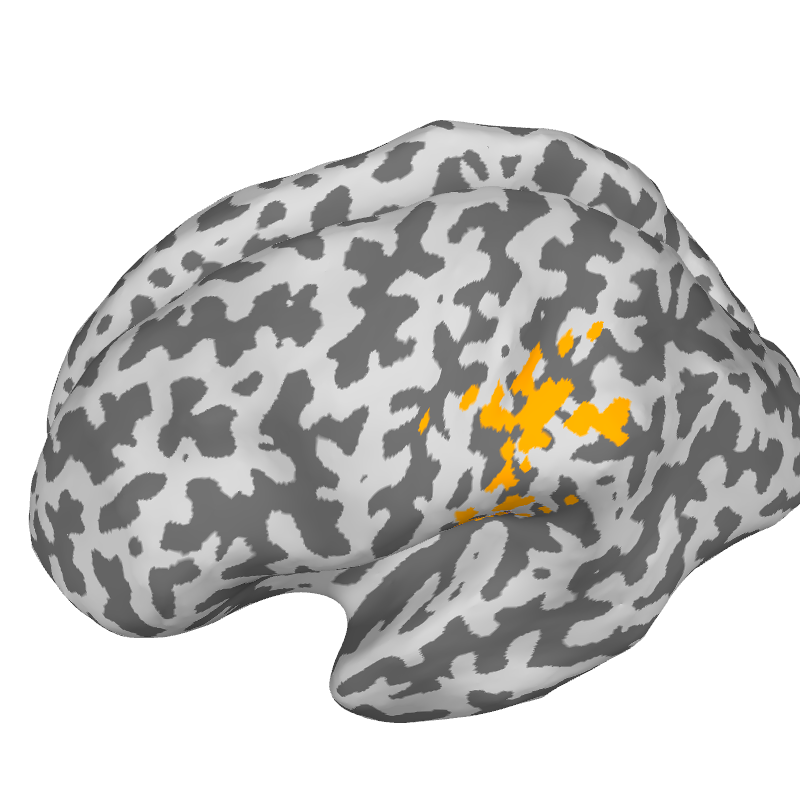
Plot 5. Spatial cohesion
Brain = get_brain_class()
knn_plot = 30
gamma_plot = [0, 0.4, 0.6, 1]
theta_plot = 0.05
cart_coord = [-0.03, 0.025, 0.08]
src = fwd['src']
V = np.concatenate((src[0]['rr'][src[0]['vertno']],
src[1]['rr'][src[1]['vertno']]), axis=0)
nvert = src[0]['nuse'] + src[1]['nuse']
idx_vert = np.argmin(np.linalg.norm(V - cart_coord, axis=1))
for gamma in gamma_plot:
target_file = string_target_file.format(
subject, knn_plot, gamma, theta_plot)
print('Loading %s'%target_file)
with open(target_file, 'rb') as aux_lf:
flame_data = pickle.load(aux_lf)
flame_labels = megicparc.store_flame_labels(flame_data, src, subject)
parcels_vector = np.zeros(nvert, dtype='int') - 1
for ir in range(flame_data['centroids']):
parcels_vector[flame_data['parcel'][ir]] = ir
sel_roi = parcels_vector[idx_vert]
index = [sel_roi + 1]
brain = megicparc.plot_flame_labels(index, flame_labels, src, subject,
subjects_dir, surf='inflated', color = [1, 0.64, 0.],
plot_region=True, plot_points=False, plot_borders=False)
brain.show_view(azimuth=173, elevation= 60)
Loading ../data/data_mne_sample/sample_flame_grad_k30_gamma0.00_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k30_gamma0.40_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k30_gamma0.60_theta0.05.pkl
Loading ../data/data_mne_sample/sample_flame_grad_k30_gamma1.00_theta0.05.pkl
Plot 7. Reduced source space on top of DK atlas
brain_redV = Brain(subject, hemi='both', surf='inflated',
background='white', subjects_dir=subjects_dir, alpha=1)
# Superimpose anatomical regions
for ir in range(len(label)):
brain_redV.add_label(label[ir], hemi=label[ir].hemi, alpha=0.9)
# Superimpose centroids
megicparc.plot_flame_centroids(flame_data, src, subject, subjects_dir,
brain_redV)
brain_redV.show_view(azimuth=170, elevation=90)
""

''
Total running time of the script: (1 minutes 38.736 seconds)